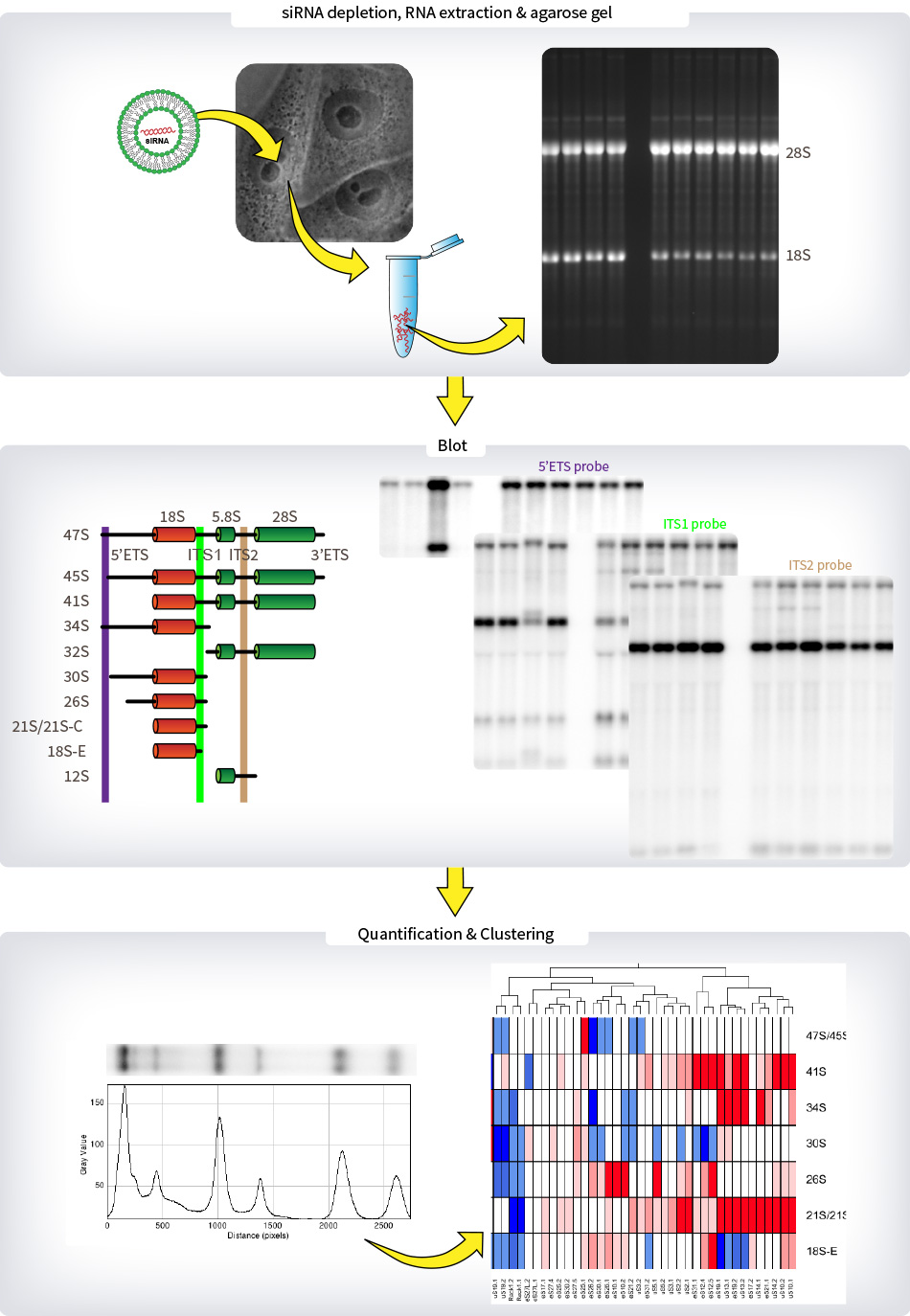
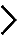Legend : METHODS – pre-rRNA Processing
Human cells were transfected with either of two distinct siRNAs specific to each of the eighty r-proteins in 6-well plates and incubated for 2 days prior to total RNA extraction and separation on a denaturing agarose gel. After transfer on a nylon membrane, three probes located in the 5’ETS, ITS-1 or ITS-2 were used to detect the various pre-rRNA precursors. These RNA species were quantified with a Phosphorimager, normalized with respect to the nontargeting control (SCR), and their abundances represented as a heatmap using the “R” software. These heatmaps are a visual representation of the logarithm of the ratio of the pre-rRNA level in the knockdown condition respective to its level in the non-targeting (Scramble, SCR) control. The calibration set used in the pre-rRNA processing analysis consists of mock-treated cells, and cells treated with a nontargeting (Scramble, SCR) siRNA, or with siRNAs specific to UTP18 or NOL9.